Biological Foundations
Using our breakthrough model of evolution, we’re aiming to establish new foundations for computational and theoretical biology.
Project Overview
In recent history science has aimed to use traditional mathematical tools to explain, predict, and understand regularities in nature. Complex phenomena were thought to require equivalently complex mathematical frameworks, usually domain-specific. A New Kind of Science introduced a different perspective: complexity in nature may arise not from elaborate mechanisms but from simple computational rules operating at all levels. More recently, the Wolfram Physics project took this idea further, suggesting that even the structure of space-time itself is the consequence of the limiting behavior of the Ruliad. We believe that, in much the same spirit, all of these concepts can be brought into biology—where the hope is that underlying computational principles may help reveal new, unifying ways of understanding life.
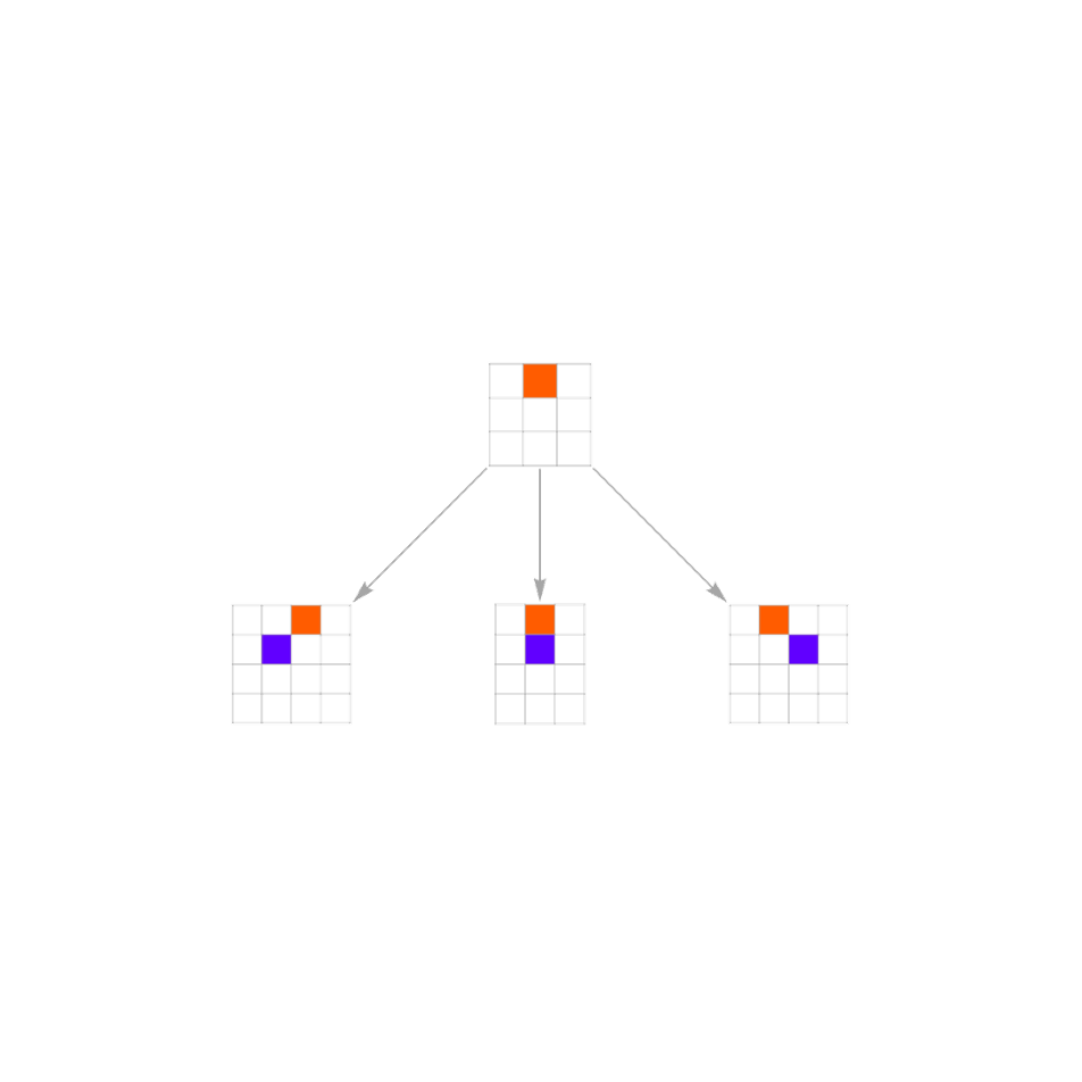
Simple Rules → Complex Behavior
Wolfram Biology Approach
Using idealized models of genetics and development, we can recover fundamental properties of living systems. Once these models are expressed in simple computational terms, it becomes clear that many biological patterns follow from general underlying rules rather than specific biological mechanisms. By abstracting away details—like specific genes, cell types, or species—we can make statements that apply across organisms and environments. As in the study of simple programs, complexity emerges not from intricate parts assembled together, but from simple rules that unfold over developmental and evolutionary timescales. This perspective points towards the existence of general unifying principles of living systems that can be found through the systematic study of biological rules.
Progress and Future Directions
Our work so far suggests that many of the intricate features of living systems can be reproduced and most importantly, understood, by studying simple computational rules. We’ve shown that patterns of growth and evolution can emerge from rules much like the ones studied in A New Kind of Science. This shift in perspective opens a new path for biology: instead of starting from the intricacies of biochemical, genetic and cellular complexity, we can start from fundamental principles expressed as simple rules. Then, we observe which life-like behavior emerges from these principles. Our early results are promising, and we are excited to uncover deeper, universal laws in living systems.
Research Output
Community Output
-
Exploring small molecule binding with interleukin proteins
Max Wang - July 11th, 2025
A method to compute and model small molecule binding with proteins in Wolfram is created along with a way to visualize residues and corresponding atom indices in a Molecule object. From the binding of the small-molecule lanraplenib to interleukin-8, lanraplenib was found to bind to the same binding site of interleukin-8 to its receptor, potentially acting as a interleukin-8 inhibitor.
-
Identifying reactive functional groups
Vikrant Chintanaboina - July 11th, 2025
This project develops functions to identify functional groups in a given molecule, aiming to improve understanding of molecular structure and reactivity.
-
Visualizing & analyzing genetic mutations causing sickle cell anemia
Jasmine Zhao - July 11th, 2025
This project uses sickle cell anemia’s E6V mutation as a case study to visualize and quantify how amino acid substitutions alter protein structure, comparing wild-type and mutant hemoglobin through 3D modeling and atomic displacement analysis.
-
Modeling DNA knot dynamics
Sarah Park - July 11th, 2025
Chromatin tends to form knots within the nucleus, which, without proper resolution by topoisomerases, can lead to cell death and mutation. However, the mechanism of this knot formation is poorly understood. Using a mechanical model, this project aims to simulate different knots along randomly moving DNA segments, and analyze the knots’ motion and size. We found that chromatin knots in this model are prone to rightward movement, size decreases, and are heavily influenced by the ratio of knot to chain length. These results provide further insight into DNA knot dynamics and can be applied to future drug development and nanotechnology.
-
Modeling tumor-immune interactions with chemotherapy via delay differential equations
Sarim Mominkhawaja - July 11th, 2025
Cancer treatment and its interaction with the human body are not well understood, making improved comprehension essential for optimizing therapies. A series of mathematical models, from tumor-immune ODEs to delay differential systems with chemotherapy and drug resistance, is developed and analyzed through simulations, biomarker assessment, and optimization. Resistance emergence, immune-tumor dynamics, and the potential superiority of adaptive dosing protocols are demonstrated, highlighting the need for dynamic, patient-specific regimens.
-
Automated insulin dosing strategy in people with type 1 diabetes
Arnay Garhyan - July 11th, 2025
Having Type 1 diabetes can become a major inconvenience in day-to-day life. Both the classical insulin injection and a hybrid closed loop insulin dosing system require extensive user input into a program to avoid extremely high or low blood sugar levels. To evaluate the capability of a two-bolus dosing system independent of any user input apart from the time of meal, the insulin-glucose response in a virtual person with Type 1 diabetes was modeled, simulated, and utilized to optimize insulin dosing. In addition, the model was used to create an interactive and educational tool to visualize the glucose and insulin levels of an individual following a meal and insulin dose. The primary result was to determine the best dosing regimen for insulin through an interactive tool for a virtual patient.
-
Genomic horizontal expansion optimization in cellular automata
Mariem El-Kady - July 11th, 2025
A cellular automaton rule is applied for optimized horizontal expansion in genomic modeling, including cases such as cells turning into infinity or similar-entropy twin rules. The rule’s behavior is examined to explore its potential for improving genome-related simulations.
-
Spatiotemporal characterization of protein molecular dynamics trajectory
Ian Asaf Muñoz Granados - July 11th, 2025
We use molecular dynamics simulation data of the CXCR7 protein. This protein is a G protein-coupled receptor (GPCRs), it scavenges chemokines and opioids, and recruit β-arrestins, so it is an atypical GPCR. For the protein helices, we extract the spatial coordinates of atoms of alpha carbon (“CA”) and monitor how their positions evolve over time using static and dynamic graphical analysis. In parallel, we compute pairwise contact distances between protein residues and analyze how these distances fluctuate throughout the simulation trajectory.
-
What shapes can organisms have? Studying biological evolution in the Wolfram evolution model
Júlia Campolim - July 11th, 2025
Due to computational irreducibility, predicting behaviour of cellular automata is a difficult, almost impossible task. However, can we induce the behaviour of a cellular automaton? More specifically, can we control its shape? In order to answer these questions, an experiment was conducted to evolve various cellular automata configurations to a specific shape, considering cellular automata boundary as its fitness function.
-
Adaptative Evolution in Totalistic Cellular Automaton
Erick Jesse Angeles Lopez - July 11th, 2025
In biology there are patterns of life that grow infinitely and uncontrollably (such as tumors), other that die out immediately under certain conditions and some other that persists for only a finite number of steps. Interestingly, some cellular automaton rules exhibit the similar behaviour. This work explore such rules within the context of one-dimensional totalistic cellular automata, focusing of those that generate finite life when starting from a single cell active.
Live Science Recordings
Associated Researchers
This research was, in part, funded by the U.S. Government.
The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the U.S. Government.